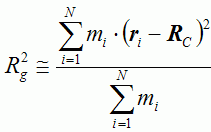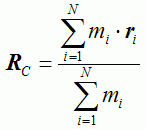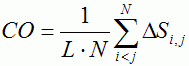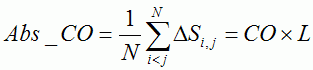
Table 1. Characteristics of proteins with multi-state kinetics | ||||||||||||||||
| Protein (or domain) name | PDB entry | fragment | Scop class | Number of amino acid residues | ln(kf) in water | Molecular Volume V, ų | Molecular Surface S, Ų | Accessible Volume V, ų | Accessible Surface S, Ų | Radius of gyration Rg, Å1 | Rg(V) of ideal ball, Å | Rg Rg(Vmol) | S(Vmol) | Relative CO2 | AbsCO3 | |
| 1 | En-HD | 1enh | A:3-56 | a | 54 | 10.5 | 7 969 | 3 854 | 13 121 | 3 788 | 10.42 | 9.60 | 1.08 | 2.0 | 13.63 | 736.2 |
| 2 | 434 Cro | 2cro | A:-1-63 | a | 65 | 3.7 | 9 010 | 4 291 | 14 836 | 4 041 | 10.65 | 10.00 | 1.06 | 2.1 | 11.21 | 728.4 |
| 3 | Ubiquitin | 1ubq | A:1-76 | d | 76 | 7.3 | 10 502 | 5 115 | 17 104 | 4 622 | 11.79 | 10.52 | 1.11 | 2.2 | 15.10 | 1 147.5 |
| 4 | Im7 | 1ayi | A:2-86 | a | 85 | 7.2 | 11 318 | 5 563 | 18 848 | 5 121 | 12.40 | 10.79 | 1.14 | 2.3 | 10.14 | 862.1 |
| 5 | HypF-N | 1gxt | A:4-91 | d | 88 | 4.4 | 12 174 | 5 329 | 19 122 | 4 925 | 12.30 | 11.05 | 1.11 | 2.1 | 20.89 | 1 838.3 |
| 6 | TI I27 | 1tit | A:1-89 | b | 89 | 3.6 | 11 734 | 6 331 | 20 254 | 5 529 | 13.16 | 10.92 | 1.20 | 2.5 | 17.80 | 1 584.1 |
| 7 | Barstar | 1brs | F:1-89 | c | 89 | 3.4 | 11 963 | 5 501 | 18 877 | 4 677 | 12.01 | 10.99 | 1.09 | 2.2 | 11.84 | 1 053.7 |
| 8 | suc1 | — | 1-96 | d | 96 | 4.2 | 14 000 | 6 440 | 22 567 | 5 879 | 13.37 | 11.58 | 1.15 | 2.3 | 13.01 | 1 249.1 |
| 9 | CD2.D1 | 1hng | A:2-96 | b | 95 | 1.8 | 12 932 | 6 244 | 20 910 | 5 500 | 13.14 | 11.28 | 1.16 | 2.3 | 16.87 | 1 602.8 |
| 10 | Barnase | 1bni | A:3-110 | d | 108 | 2.6 | 14 442 | 6 122 | 22 595 | 5 630 | 13.45 | 11.70 | 1.15 | 2.1 | 11.35 | 1 225.9 |
| 11 | Villin 14T3 | 2vik | A:1-126 | d | 126 | 5.0 | 16 355 | 7 581 | 26 777 | 7 205 | 13.92 | 12.20 | 1.14 | 2.4 | 12.26 | 1 544.5 |
| 12 | p16 | 2a5e | A:10-136 | a | 127 | 3.5 | 15 975 | 7 575 | 32 061 | 8 726 | 17.81 | 12.10 | 1.15 | 2.8 | 5.33 | 831.5 |
| 13 | ILBP | 1eal | A:1-127 | b | 127 | 1.3 | 17 163 | 8 631 | 28 127 | 6 806 | 14.18 | 12.40 | 1.14 | 2.7 | 12.40 | 1 574.9 |
| 14 | CheY | 3chy | A:2-129 | c | 128 | 1.0 | 17 009 | 7 671 | 26 504 | 6 216 | 13.43 | 12.36 | 1.08 | 2.4 | 8.71 | 1 115.0 |
| 15 | IFABP | 1ifc | A:1-131 | b | 131 | 3.4 | 18 208 | 8 250 | 28 406 | 6 571 | 14.00 | 12.64 | 1.10 | 2.5 | 13.52 | 1 771.0 |
| 16 | CRBPII | 1opa | A:1-133 | b | 133 | 1.4 | 18 853 | 8 423 | 29 463 | 6 860 | 14.30 | 12.79 | 1.12 | 2.5 | 14.02 | 1 865.2 |
| 17 | CRABPI | 1cbi | A:1-136 | b | 136 | -3.2 | 18 579 | 8 479 | 29 627 | 7 316 | 14.54 | 12.73 | 1.14 | 2.5 | 13.81 | 1 877.7 |
| 18 | apoMb | 1a6n | A:1-151 | a | 151 | 1.1 | 20 456 | 9 089 | 32 162 | 7 481 | 15.14 | 13.14 | 1.15 | 2.5 | 8.38 | 1 265.7 |
| 19 | GroEL(191-345) | 1aon | A:191-345 | c | 155 | -1.5 | 18 394 | 8 770 | 30 176 | 7 226 | 14.40 | 12.68 | 1.13 | 2.6 | 13.66 | 2 116.9 |
| 20 | RNase HI | 2rn2 | A:1-155 | c | 155 | 0.1 | 21 290 | 9 583 | 33 542 | 8 172 | 15.53 | 13.32 | 1.16 | 2.6 | 12.43 | 1 927.1 |
| 21 | DHFR | 1ra9 | A:1-159 | c | 159 | -3.2 | 21 753 | 9 805 | 34 051 | 8 377 | 15.57 | 13.41 | 1.16 | 2.6 | 14.03 | 2 231.4 |
| 22 | T4L | 2lzm | A:1-164 | d | 164 | 4.1 | 22 486 | 9 797 | 34 732 | 8 217 | 16.53 | 13.56 | 1.22 | 2.6 | 7.07 | 1 160.3 |
| 23 | N-terminal domain from PGK | 1php | A:1-175 | c | 175 | 2.3 | 23 769 | 10 587 | 36 320 | 8 255 | 15.18 | 13.82 | 1.10 | 2.6 | 11.55 | 2 020.9 |
| 24 | C-terminal domain from PGK | 1php | A:176-394 | c | 219 | -3.5 | 28 715 | 12 851 | 44 144 | 10 014 | 17.30 | 14.71 | 1.17 | 2.8 | 7.96 | 1 742.5 |
| 25 | Tryptophan syntase α subunit | 1qop | A:1-267 | c | 267 | -2.5 | 34 239 | 14 124 | 51 202 | 10 687 | 17.33 | 15.60 | 1.10 | 2.8 | 8.35 | 2 228.2 |
| 26 | Tryptophan syntase β subunit | 1qop | B:2-391 | c | 390 | -6.9 | 50 537 | 19 088 | 72 427 | 13 812 | 19.72 | 17.77 | 1.11 | 2.9 | 8.34 | 3 253.4 |
Table 2. Characteristics of proteins with two-state kinetics | ||||||||||||||||
| Protein (or domain) name | PDB entry | fragment | Scop class | Number of amino acid residues | ln(kf) in water | Molecular Volume V, ų | Molecular Surface S, Ų | Accessible Volume V, ų | Accessible Surface S, Ų | Radius of gyration Rg, Å1 | Rg(V) of ideal ball, Å | Rg Rg(Vmol) | S(Vmol) | Relative CO2 | AbsCO | |
| 27 | β-hairpin | 1pgb | A:41-56 | - | 16 | 12.0 | 2 172 | 1 305 | 4 385 | 1 714 | 8.36 | 6.22 | 1.32 | 1.6 | 25.83 | 413.2 |
| 28 | α-helix | — | 1-21 | - | 21 | 15.5 | 1 694 | 1 086 | 3 976 | 1 699 | 9.42 | 5.73 | 1.65 | 1.6 | 10.42 | 218.9 |
| 29 | Trp-cage protein | 1l2y | A:1-20 | a | 20 | 20.5 | 2 577 | 1 468 | 4 984 | 1 828 | 7.25 | 6.59 | 1.10 | 1.6 | 18.70 | 374.1 |
| 30 | BBA5 | 1t8j | A:1-23 | d | 23 | 11.8 | 3 279 | 2 035 | 6 508 | 2 433 | 8.86 | 7.14 | 1.23 | 1.9 | 12.78 | 293.9 |
| 31 | Pin WW domain | 1pin | A:6-39 | b | 34 | 9.5 | 4 793 | 2 387 | 8 415 | 2 813 | 9.50 | 8.10 | 1.17 | 1.7 | 19.04 | 647.2 |
| 32 | HP36 | 1vii | A:41-76 | a | 36 | 9.4 | 4 847 | 2 680 | 9 103 | 3 044 | 9.43 | 8.13 | 1.15 | 1.9 | 11.19 | 403.0 |
| 33 | WW prototype | 1e0m | A:1-37 | b | 37 | 8.9 | 5 036 | 2 641 | 9 115 | 3 119 | 9.66 | 8.24 | 1.16 | 1.9 | 17.81 | 658.9 |
| 34 | FBP28 | 1e0l | A:1-37 | b | 37 | 10.6 | 5 097 | 2 773 | 9 265 | 3 082 | 9.85 | 8.27 | 1.19 | 1.9 | 16.29 | 602.7 |
| 35 | YAP65 | 1jmq | A:5-44 | b | 40 | 8.4 | 5 419 | 2 800 | 9 609 | 3 125 | 9.81 | 8.44 | 1.16 | 1.9 | 22.50 | 900.1 |
| 36 | psbd41 | 2pdd | A:3-43 | a | 41 | 9.8 | 5 205 | 3 106 | 9 851 | 3 311 | 9.43 | 8.33 | 1.13 | 2.1 | 11.45 | 469.6 |
| 37 | Albumin-binding domain of protein PAB | 1prb | A:7-53 | a | 47 | 13.8 | 6 012 | 3 224 | 10 752 | 3 414 | 9.73 | 8.74 | 1.11 | 2.0 | 12.34 | 580.0 |
| 38 | c-Myb | 1gv2 | A:144-191 | a | 47 | 8.7 | 6 759 | 3 388 | 11 481 | 3 435 | 9.78 | 9.09 | 1.07 | 2.0 | 12.30 | 578.1 |
| 39 | TRF1 | 1ba5 | A:5-53 | a | 49 | 5.9 | 7 300 | 3 582 | 12 468 | 3 651 | 10.04 | 9.32 | 1.07 | 2.0 | 14.52 | 711.6 |
| 40 | Src SH3 | 1rlq | C:9-64 | b | 56 | 4.4 | 7 499 | 3 894 | 13 123 | 4 001 | 10.47 | 9.41 | 1.11 | 2.1 | 20.29 | 1 136.3 |
| 41 | NTL9 | 1div | A:1-56 | d | 56 | 6.6 | 7 434 | 3 712 | 12 922 | 3 925 | 11.61 | 9.38 | 1.24 | 2.0 | 12.66 | 709.0 |
| 42 | Protein G | 3gb1 | A:1-56 | d | 56 | 6.3 | 7 370 | 3 957 | 12 853 | 3 779 | 10.71 | 9.35 | 1.14 | 2.2 | 16.52 | 925.3 |
| 43 | Spectrin SH3 | 1shg | A:6-62 | b | 57 | 1.1 | 8 050 | 3 868 | 13 387 | 3 801 | 10.30 | 9.63 | 1.07 | 2.0 | 19.10 | 1 088.9 |
| 44 | Fyn SH3 | 1avz | A:85-141 | b | 57 | 4.9 | 8 169 | 4 532 | 13 030 | 3 715 | 10.24 | 9.68 | 1.19 | 2.0 | 19.62 | 1 118.4 |
| 45 | Abp1 SH3 | 1jo8 | A:1-58 | b | 58 | 2.5 | 7 936 | 3 602 | 12 951 | 3 618 | 10.25 | 9.58 | 1.06 | 1.9 | 19.39 | 1 124.7 |
| 46 | RAP1 | 1fex | A:1-59 | a | 59 | 8.2 | 7 841 | 4 222 | 13 872 | 4 093 | 10.56 | 9.55 | 1.10 | 2.2 | 13.04 | 769.6 |
| 47 | BdpA | 1bdd | A:1-60 | a | 60 | 11.7 | 7 789 | 4 140 | 13 797 | 4 394 | 12.09 | 9.53 | 1.26 | 2.2 | 8.70 | 522.1 |
| 48 | Protein L | 2ptl | A:18-77 | d | 60 | 4.1 | 7 634 | 3 768 | 13 057 | 3 765 | 11.06 | 9.46 | 1.17 | 2.0 | 18.49 | 1 109.3 |
| 49 | Sso7d | 1c8c | A:2-64 | d | 63 | 7.0 | 8 520 | 4 341 | 15 400 | 4 721 | 11.26 | 9.81 | 1.13 | 2.2 | 12.67 | 798.1 |
| 50 | Ci2 | 2ci2 | I:20-83 | d | 64 | 5.8 | 8 896 | 4 343 | 15 150 | 4 325 | 11.25 | 9.96 | 1.13 | 2.1 | 15.65 | 1 001.7 |
| 51 | CspB | 1c9o | A:1-66 | b | 66 | 7.2 | 8 902 | 4 363 | 14 886 | 4 313 | 11.06 | 9.96 | 1.10 | 2.1 | 16.90 | 1 115.4 |
| 52 | CspB | 1g6p | A:1-66 | b | 66 | 6.3 | 8 801 | 4 485 | 15 164 | 4 551 | 11.01 | 9.92 | 1.11 | 2.2 | 17.71 | 1 168.7 |
| 53 | CspB | 1csp | A:1-67 | b | 67 | 6.5 | 8 669 | 4 327 | 14 682 | 4 293 | 10.94 | 9.87 | 1.10 | 2.1 | 16.39 | 1 098.5 |
| 54 | EC298 | 1jyg | A:1-69 | a | 69 | 9.1 | 9 585 | 4 722 | 16 004 | 4 478 | 11.31 | 10.21 | 1.10 | 2.2 | 12.07 | 832.6 |
| 55 | PsaE | 1psf | A:1-69 | b | 69 | 3.2 | 8 994 | 4 750 | 15 732 | 4 863 | 12.32 | 9.99 | 1.23 | 2.3 | 17.03 | 1 174.8 |
| 56 | CspA | 1mjc | A:2-70 | b | 69 | 5.3 | 8 761 | 4 185 | 14 616 | 4 124 | 10.82 | 9.91 | 1.09 | 2.0 | 15.96 | 1 101.5 |
| 57 | ADAh2 | 1o6x | A:9-79 | d | 71 | 6.8 | 9 425 | 4 814 | 19 527 | 5 963 | 14.70 | 10.15 | 1.10 | 2.5 | 16.34 | 1 323.5 |
| 58 | α3D | 2a3d | A:1-73 | a | 73 | 12.2 | 9 635 | 5 377 | 17 092 | 5 022 | 13.05 | 10.23 | 1.27 | 2.5 | 9.54 | 696.2 |
| 59 | GW1 | 1m9s | A:391-466 | b | 76 | 4.0 | 10 507 | 5 332 | 17 549 | 4 911 | 11.83 | 10.52 | 1.12 | 2.3 | 18.27 | 1 388.2 |
| 60 | raf RBD | 1rfa | A:55-132 | d | 78 | 8.4 | 10 365 | 4 819 | 16 972 | 4 562 | 11.49 | 10.48 | 1.09 | 2.1 | 16.17 | 1 261.4 |
| 61 | λ-repressor | 1lmb | 3:6-85 | a | 80 | 10.4 | 10 587 | 5 128 | 17 333 | 4 662 | 11.88 | 10.55 | 1.12 | 2.2 | 9.37 | 749.6 |
| 62 | Im9 | 1imq | A:2-86 | a | 85 | 7.3 | 10 548 | 5 072 | 17 701 | 5 180 | 12.05 | 10.54 | 1.14 | 2.2 | 12.28 | 1 043.8 |
| 63 | HPr | 1poh | A:1-85 | d | 85 | 2.7 | 11 013 | 5 121 | 17 821 | 4 685 | 11.77 | 10.69 | 1.10 | 2.1 | 17.64 | 1 499.2 |
| 64 | ACBP | 1nti | A:1-86 | a | 86 | 7.0 | 11 434 | 5 356 | 19 209 | 5 329 | 13.16 | 10.83 | 1.21 | 2.2 | 12.37 | 1 064.2 |
| 65 | PI3-SH3 | 1pnj | A:1A-84 | b | 86 | -1.0 | 11 671 | 6 511 | 20 251 | 5 634 | 12.85 | 10.90 | 1.17 | 2.6 | 16.13 | 1 387.1 |
| 66 | TNfn3 | 1ten | A:803-891 | b | 89 | 1.1 | 11 931 | 5 568 | 19 105 | 4 964 | 13.12 | 10.98 | 1.19 | 2.2 | 17.35 | 1 544.4 |
| 67 | 9FNIII | 1fnf | A:1326-1415 | b | 90 | -0.9 | 11 832 | 5 141 | 18 989 | 5 068 | 12.92 | 10.95 | 1.18 | 2.1 | 18.09 | 1 628.0 |
| 68 | CTL9 | 1div | A:58-149 | d | 92 | 3.3 | 12 125 | 5 828 | 19 895 | 5 482 | 14.18 | 11.04 | 1.28 | 2.3 | 13.81 | 1 270.7 |
| 69 | TWIg18’ | 1wit | A:1-93 | b | 93 | 0.4 | 11 478 | 5 495 | 19 471 | 5 544 | 13.19 | 10.84 | 1.21 | 2.2 | 20.32 | 1 890.0 |
| 70 | FNfn10 | 1fnf | A:1416-1509 | b | 94 | 5.5 | 11 794 | 5 402 | 19 342 | 5 346 | 13.07 | 10.94 | 1.19 | 2.1 | 16.51 | 1 551.6 |
| 71 | L23 | 1n88 | A:1-96 | d | 96 | 2.0 | 12 567 | 6 114 | 21 478 | 6 213 | 14.30 | 11.17 | 1.28 | 2.3 | 14.09 | 1 352.2 |
| 72 | U1A | 1urn | A:2-97 | d | 96 | 4.6 | 13 324 | 5 955 | 21 469 | 5 593 | 12.59 | 11.39 | 1.10 | 2.2 | 16.90 | 1 622.8 |
| 73 | S6 | 1ris | A:1-97 | d | 97 | 6.1 | 14 142 | 6 822 | 22 661 | 5 814 | 13.55 | 11.62 | 1.16 | 2.4 | 18.95 | 1 837.9 |
| 74 | CT AcP | 2acy | A:1-98 | d | 98 | 0.8 | 13 437 | 6 077 | 21 309 | 5 511 | 12.64 | 11.42 | 1.11 | 2.2 | 20.03 | 1 962.5 |
| 75 | mAcP | 1aps | A:1-98 | d | 98 | -1.6 | 13 206 | 5 862 | 21 067 | 5 325 | 12.86 | 11.36 | 1.13 | 2.2 | 21.20 | 2 077.5 |
| 76 | SrcSH2 | 1spr | A:2-104 | d | 103 | 8.7 | 14 238 | 6 231 | 22 455 | 5 776 | 13.20 | 11.65 | 1.13 | 2.2 | 11.65 | 1 199.6 |
| 77 | cyt b562 | 256b | A:1-106 | a | 106 | 12.3 | 14 052 | 6 764 | 22 954 | 5 959 | 14.52 | 11.60 | 1.25 | 2.4 | 7.49 | 793.5 |
| 78 | R16 | 1cun | A:7-112 | a | 110 | 4.8 | 14 745 | 7 150 | 24 475 | 6 720 | 17.20 | 11.78 | 1.46 | 2.5 | 10.35 | 1 097.2 |
| 79 | R17 | 1cun | A:113-219 | a | 107 | 3.4 | 14 226 | 6 875 | 23 828 | 6 582 | 17.24 | 11.64 | 1.48 | 2.4 | 10.22 | 1 093.5 |
| 80 | FKBP | 1fkf | A:1-107 | d | 107 | 1.6 | 14 427 | 6 443 | 22 704 | 5 783 | 13.28 | 11.70 | 1.13 | 2.3 | 17.61 | 1 884.6 |
| 81 | R15 | 1u5p | A:1662-1771 | a | 110 | 11.0 | 14 822 | 7 067 | 24 777 | 6 931 | 17.40 | 11.80 | 1.47 | 2.4 | 10.31 | 1 133.9 |
| 82 | CheW | 1k0s | A:9-151 | b | 143 | 7.4 | 18 785 | 9 206 | 31 225 | 8 243 | 14.88 | 12.77 | 1.16 | 2.7 | 13.29 | 1 901.0 |
| 83 | CyPA | 1lop | A:1-164 | b | 164 | 6.6 | 21 985 | 9 193 | 33 246 | 7 543 | 14.75 | 13.46 | 1.09 | 2.4 | 15.68 | 2 570.9 |
| 84 | VlsE | 1l8w | D:29-344 | a | 294 | 2.0 | 34 753 | 15 488 | 54 792 | 12 119 | 22.24 | 15.68 | 1.37 | 3.0 | 8.48 | 2 508.6 |